Streaming/Online vs Offline data
Neuropype can process both streaming data such as "live" data coming from an EEG headset or other device, and previously recorded data which is loaded from disk. These are often referred to as "streaming"/"online" and "offline" data, respectively (not in the Internet sense). Many of the example pipelines and documentation refer to processing streaming data which is typically the case for BCI. However, Neuropype can just as easily process recorded data. Most nodes can handle either streaming/online or recorded/offline data, though some can only handle one or the other in which case this is indicated in the documentation for that node.
Generally speaking, nodes behave the same when processing online or offline data: a Packet comes into the node, the node performs some transformation on the data in the Packet, and then passes the Packet to the next node. However, there are two important differences to bear in mind:
1) With offline/recorded data, the entire dataset is read from a file and passed along from node to node in a single Packet. This means that the pipeline typically does a single pass through each node after which it is finished (exceptions to this include when processing multiple files using the PathIterator node, in which case the pipeline would repeat for each file). Once the data is processed, the pipeline will automatically terminate.
2) With online/streaming data, data is received from a device a few samples at a time. So each Packet contains a number of samples which are passed through the pipeline from end to end. The pipeline then repeats with each new Packet. NeuroPype runs at around 20Hz by default, though very heavy computational pipelines may run more slowly. If your data sampling rate is 250Hz and the pipeline is running at 10Hz, each Packet will contain about 25 samples. Unlike with offline data, the pipeline will continue to run until it is manually stopped, even if there is no more data coming in (so that it doesn't terminate automatically in the event there is a short pause in the data).
Getting recorded data in and out
To process recorded data, you would usually start with an ImportFile node (which will call the correct importer based on the file extension; you can also call a specific import node directly which may have additional parameters specific to that file format), then connect other nodes as desired. In most cases you'll want to export the processed data and/or computed results to disk, for which you can use one of the Export nodes. Common use cases are: * ExportXDF for exporting processed signal/timeseries data to XDF format (an excellent choice for multimodal signal data; XDF import/export libraries are available for Python and MATLAB); * ExportH5 for exporting computed measures which may have additional axes (i.e., Feature, Frequency, etc.) to HDF5 format (a highly flexible format for multi-dimensional data which can be opened in many other applications, including MATLAB); * ExportCSV for exporting computed results to CSV (primarily useful for 1D or 2D data)
Writing streaming data to disk
You can also write online/streaming data to disk using the Record nodes (RecordToXDF, RecordToCSV, RecordToREC) instead of the Export nodes. These nodes write data to a file in realtime (so no data is lost in the event of an unexpected shutdown). RecordToXDF and RecordToCSV write data continuously to XDF and CSV files, respectively. RecordToREC records data to disk in Neuropype's native format, and so provides a lossless way of writing data to disk that can later be imported and played back in a pipeline using the PlayREC node. RecordToREC can be used to record either streaming data (multiple packets) or offline data (a single packet).
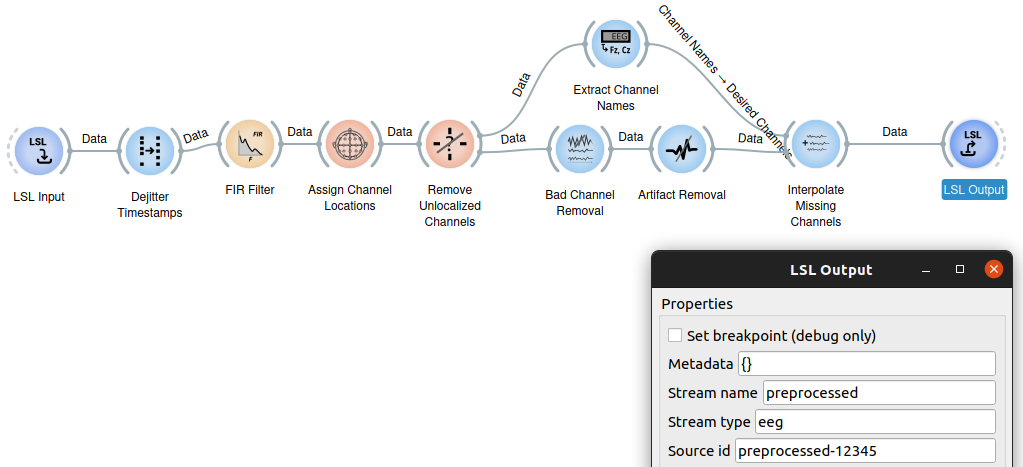
Streaming recorded data ("playback")
Neuropype also makes it easy to simulate a live environment by importing a recorded file and "playing it back", essentially streaming it as if it were streaming data coming from an LSL stream. For this, the StreamData node is used, which can stream data either in a mode that mimics the slight jitter one would expect from a live device, or in a deterministic mode with an exact number of samples per packet (usually for testing or special cases). You'll find this approach used in many of the example pipelines included with Neuropype. (In many of those example pipelines, a file is imported with ImportXDF and streamed with StreamData over an LSL stream created with LSLOutput, that same LSL stream is read in with an LSLInput node and passed on to other nodes for processing. The trip through LSL is provided for example purposes but that can be skipped and you can directly connect the StreamData node to the processing nodes (i.e., FIRFilter, etc.).
Streaming recorded data is primarily useful for real-time visualizations or perhaps for creating an LSL stream consumed by another application (it's also great for testing a live setup without having to have the device on). For data analysis, it's better to process recorded data in offline (non-streaming) mode (that is, without using the StreamData node). Not only it is much faster, but it also allows nodes which calibrate on the data, such as ArtifactRemoval, BadChannelRemoval, etc., to "see" the entire dataset rather than only a fraction of it, increasing their accuracy.
Buffering streaming data
In some cases you may want to buffer streaming data to then be processed as a single chunk (in a sense, converting "online" data to "offline" data, the opposite of StreamData). One way to do that is to use the AccumulateCalibrationData which has options to determine how much data to buffer (based on event markers or times), and whether to emit or drop the data before or after the buffered block, or to drop the buffered block itself. Despite the name, this node is not limited to calibration purposes. For example, you could use it to compute the average for a baseline period, or perform a computation only after the baseline, etc.
In the example snippet below, the AccumulateCalibrationData node is used twice, with different settings, one to compute an average of a 30 seconds baseline period and then continuously retransmit that same value using the HoldLastPacket node. In the other case, the AccumulateCalibrationData node is used to drop the baseline and calculate a histogram on the data after the first 30 seconds. In this example, both instances of the AccumulateCalibrationData have ParameterPort nodes which can be set over the Neuropype API, to determine the baseline begin and end times.
Handling streaming and offline data in nodes
A stream will have a is_streaming flag to indicate to nodes that the data is streaming/online. If the data is offline, that flag will not be present (or set to False). Input nodes, such as LSLInput or ImportFile, are responsible to set the flag. Certain nodes, such as StreamData may change the data from offline to streaming and set that flag accordingly.
If you write a node that only operates on either offline or streaming data, or which needs to handle both types different (i.e., buffering in the case of streaming data), use enumerate_chunks(with_flags=Flags.is_streaming) to only select streams with streaming data (or conversely without_flags to only select offline streams). If you're writing an input node, be sure your node sets that flag appropriately.